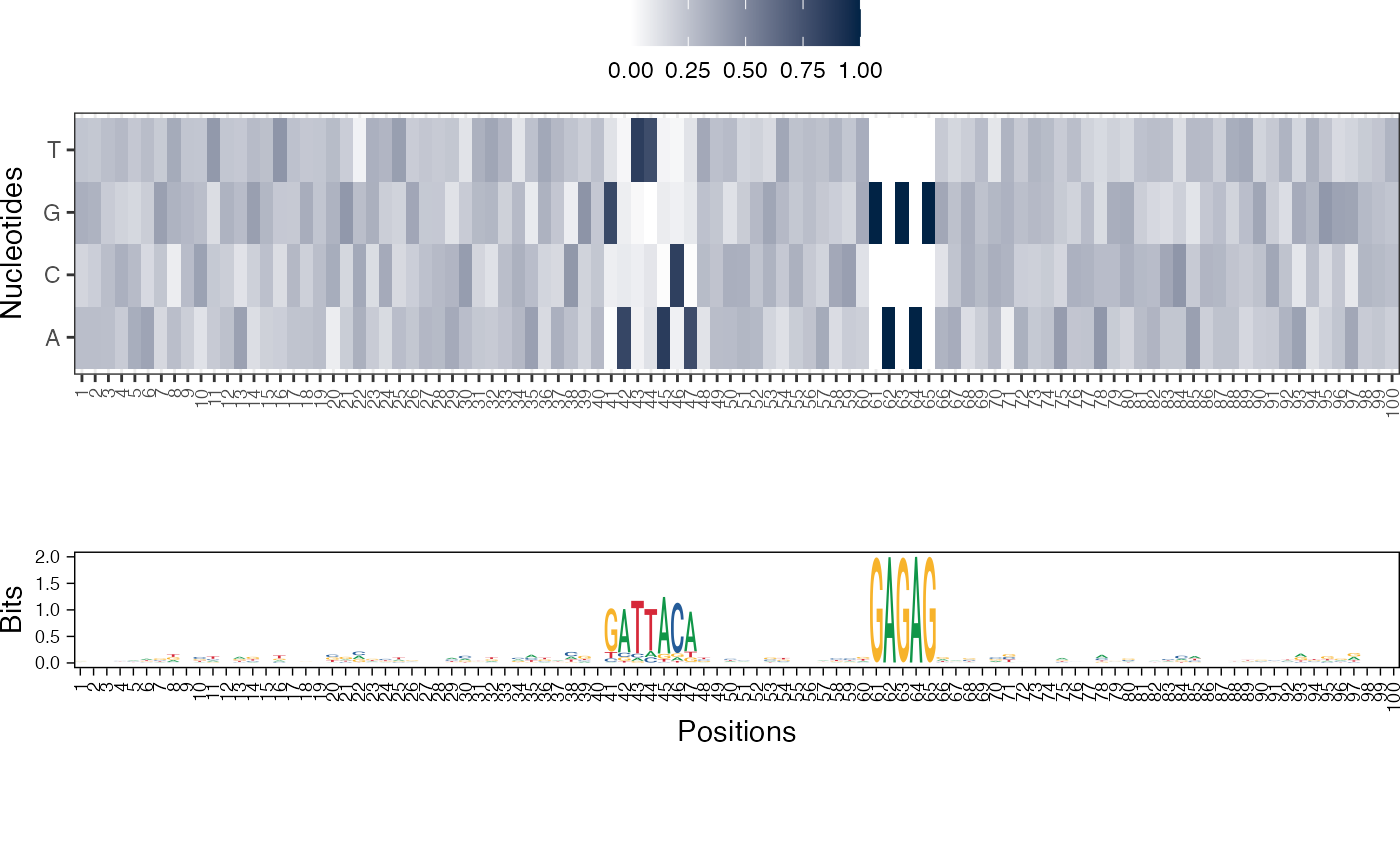
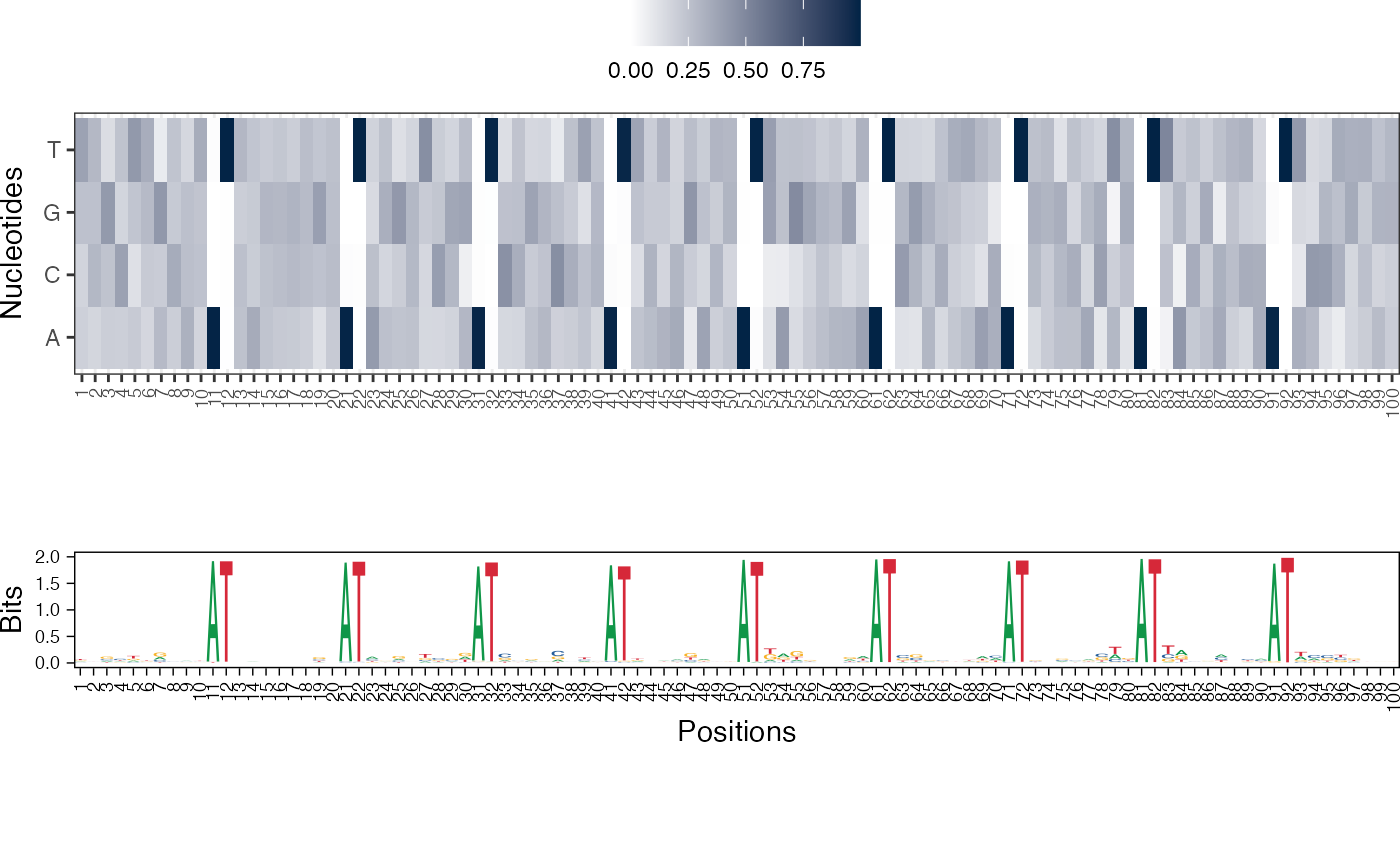
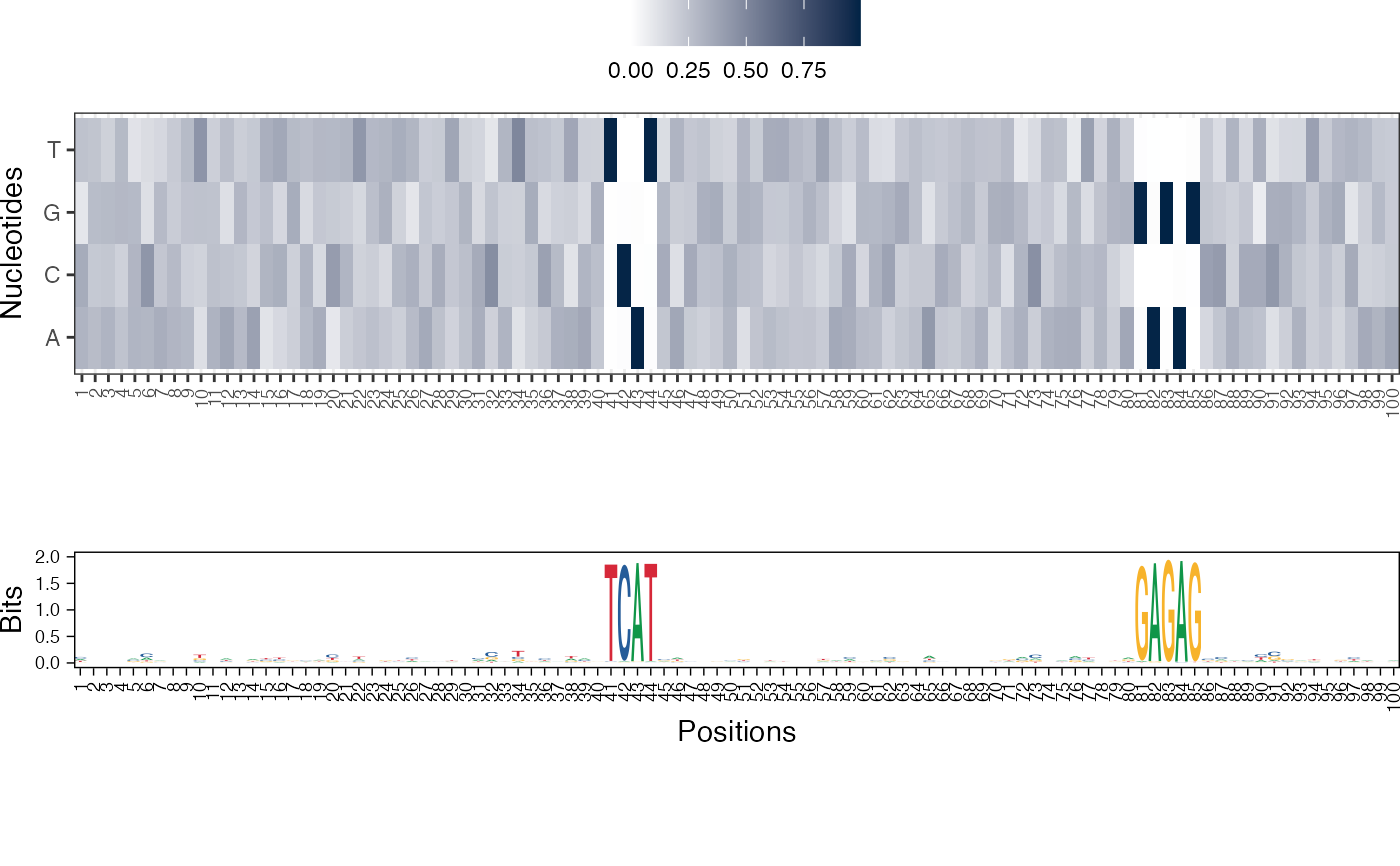
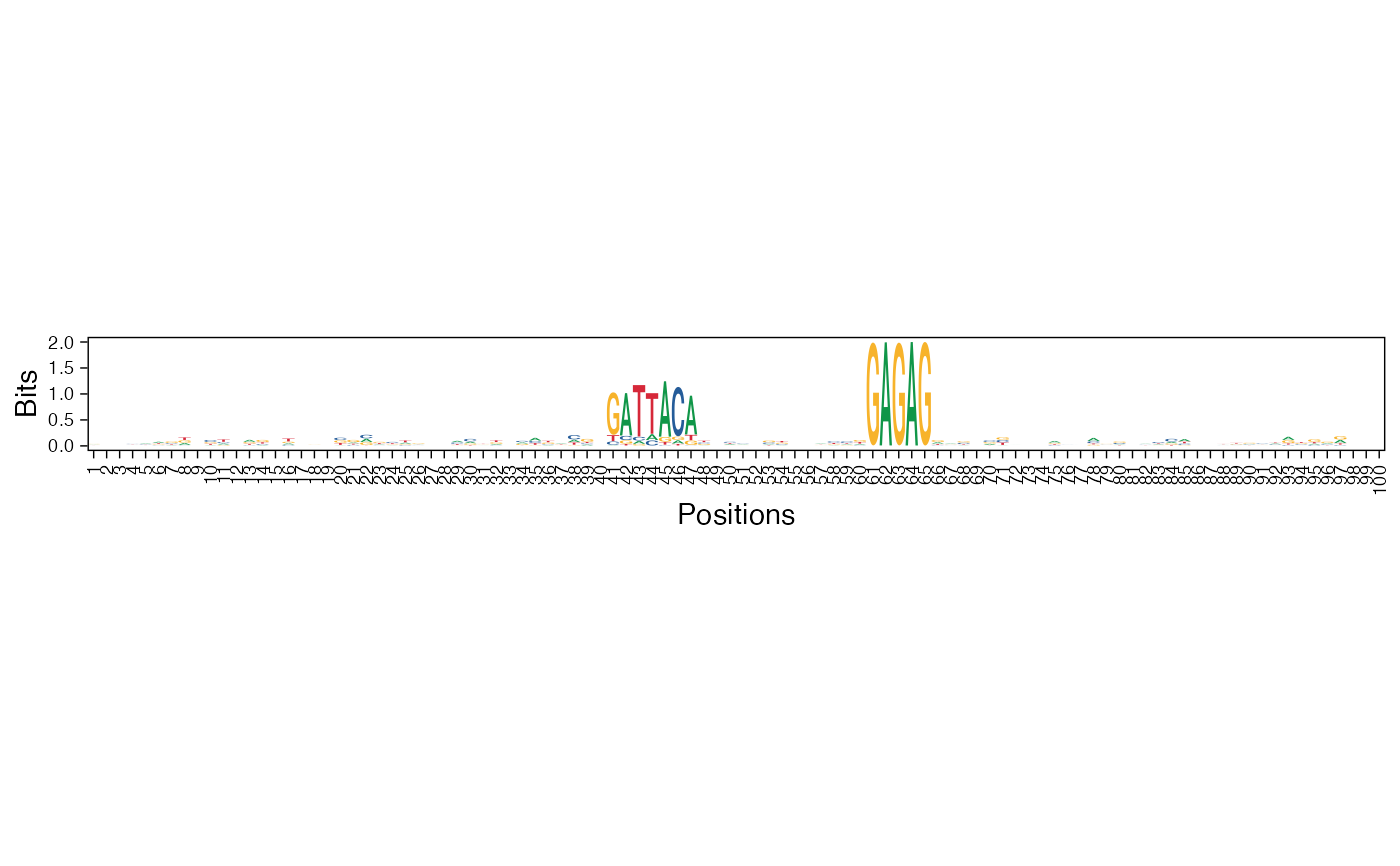
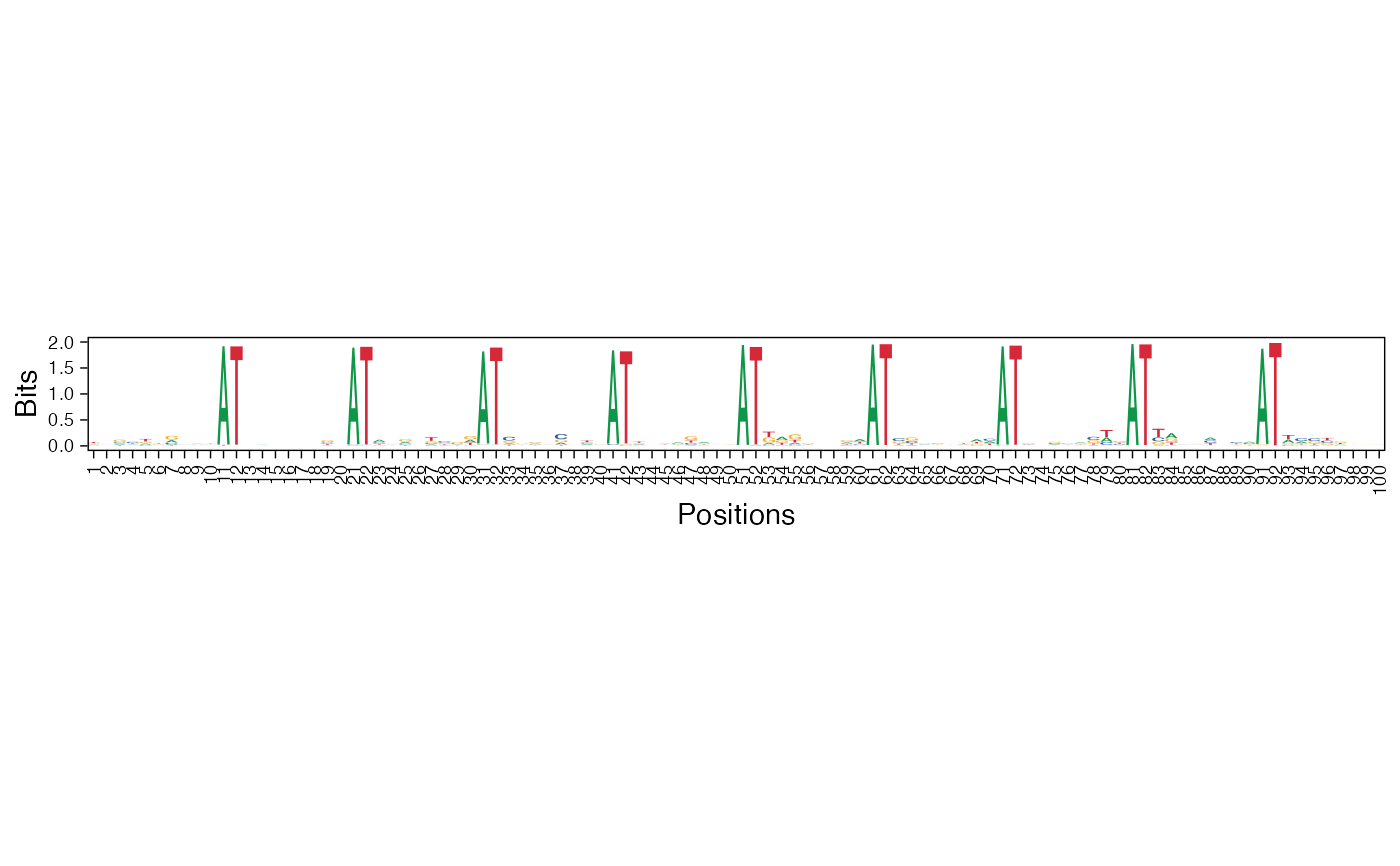
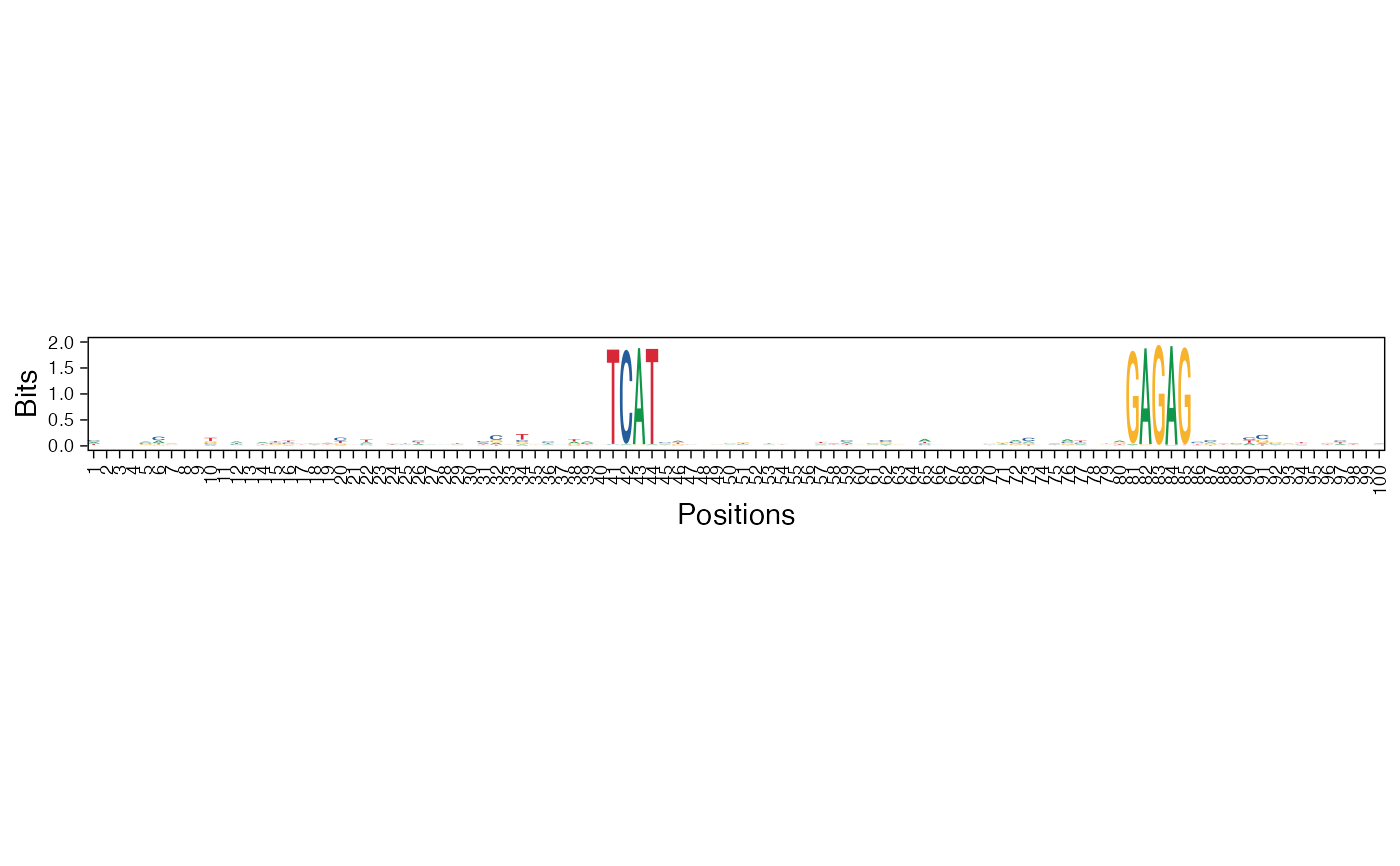
Visualize the NMF basis vectors in a paired heatmap and sequence logo plot
Source:R/viz_basis_vectors.R
viz_bas_vec_heatmap_seqlogo.RdThe given features matrix is visualized as a heatmap followed by a sequence logo where the positions are aligned for better visualization.
viz_bas_vec_heatmap_seqlogo( feat_mat, method = "bits", pos_lab = NULL, add_pseudo_counts = FALSE, pdf_name = NULL, sinuc_or_dinuc = "sinuc", fixed_coord = FALSE ) viz_bas_vec_seqlogo( feat_mat, method = "bits", pos_lab = NULL, add_pseudo_counts = FALSE, pdf_name = NULL, sinuc_or_dinuc = "sinuc", fixed_coord = FALSE ) viz_bas_vec_heatmap( feat_mat, pos_lab = NULL, add_pseudo_counts = FALSE, pdf_name = NULL, sinuc_or_dinuc = "sinuc", fixed_coord = FALSE )
Arguments
| feat_mat | The features matrix (basis vectors matrix) from archR. |
|---|---|
| method | For |
| pos_lab | Labels for sequence positions, should be of same length as that of the sequences. Default value is NULL, when the positions are labeled from 1 to the length of the sequences. |
| add_pseudo_counts | Logical, taking values TRUE or FALSE, default set to FALSE. Setting it to TRUE will enable adding pseudo-counts to the features matrix. |
| pdf_name | Name of the file which will be saved as PDF (also provide the extension). |
| sinuc_or_dinuc | "sinuc" or "dinuc" for choosing between mono- and dinucleotide profiles respectively. |
| fixed_coord | Set this to TRUE to use a fixed aspect ratio for the plot. Default is FALSE. |
Value
nothing
Functions
viz_bas_vec_seqlogo: Visualize the NMF basis vectors as a sequence logoviz_bas_vec_heatmap: Visualize the NMF basis vectors as a heatmap
See also
Other visualization functions:
plot_ggheatmap(),
plot_ggseqlogo(),
viz_seqs_acgt_mat_from_seqs()
Examples
res <- readRDS(system.file("extdata", "example_archRresult.rds", package = "archR", mustWork = TRUE)) viz_bas_vec_heatmap_seqlogo(feat_mat = get_clBasVec_m(res,iter=1), sinuc_or_dinuc = "dinuc", fixed_coord = TRUE)#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> #>#>#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> #>#>#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> #>#>viz_bas_vec_seqlogo(feat_mat = get_clBasVec_m(res,iter=1), sinuc_or_dinuc = "dinuc", fixed_coord = TRUE)#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> #>#>#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> #>#>#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> #>#># Visualizing basis vector for a single cluster viz_bas_vec_heatmap(feat_mat = as.matrix(get_clBasVec_m(res,iter=1)[,3]), sinuc_or_dinuc = "dinuc", fixed_coord = TRUE)