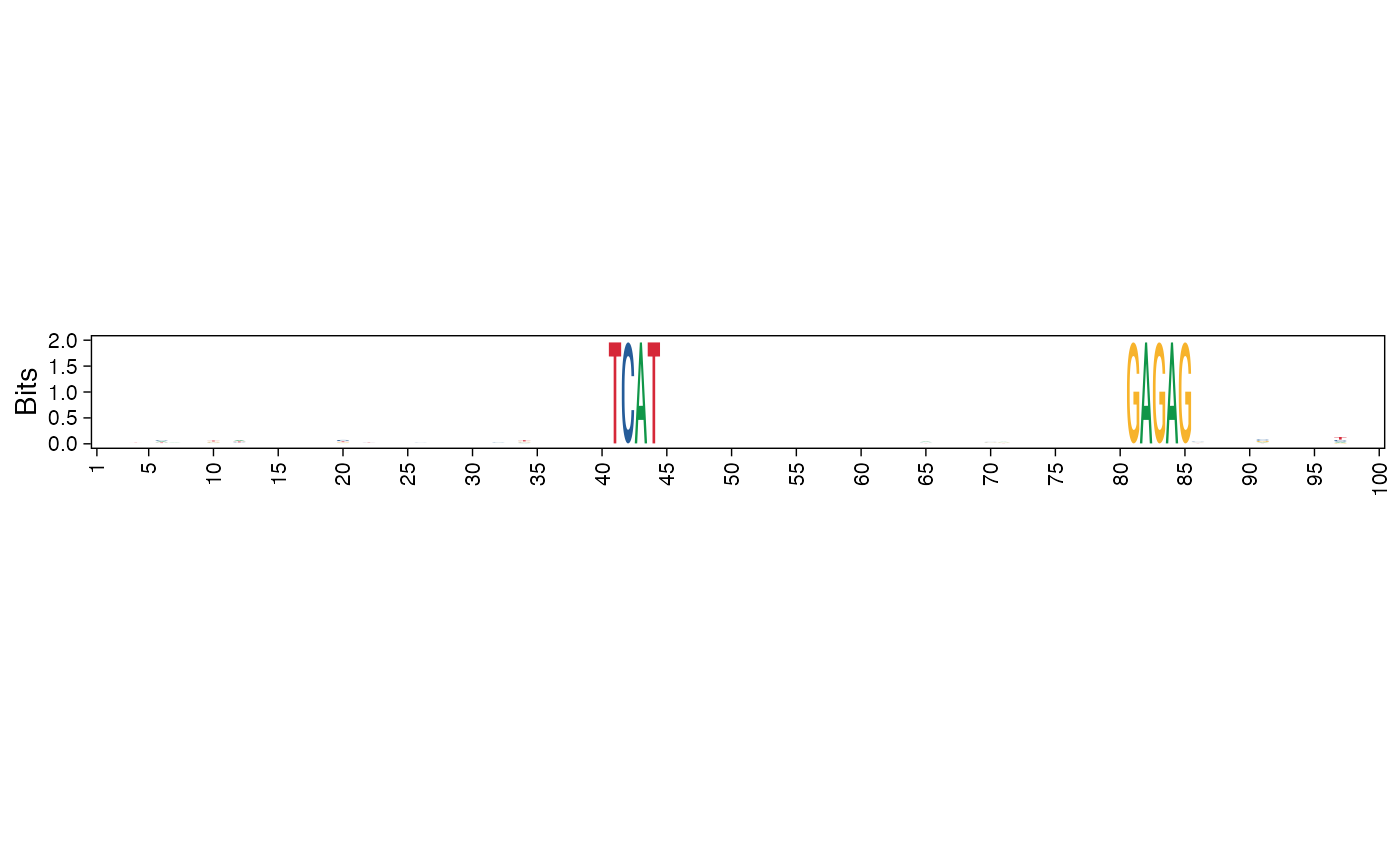
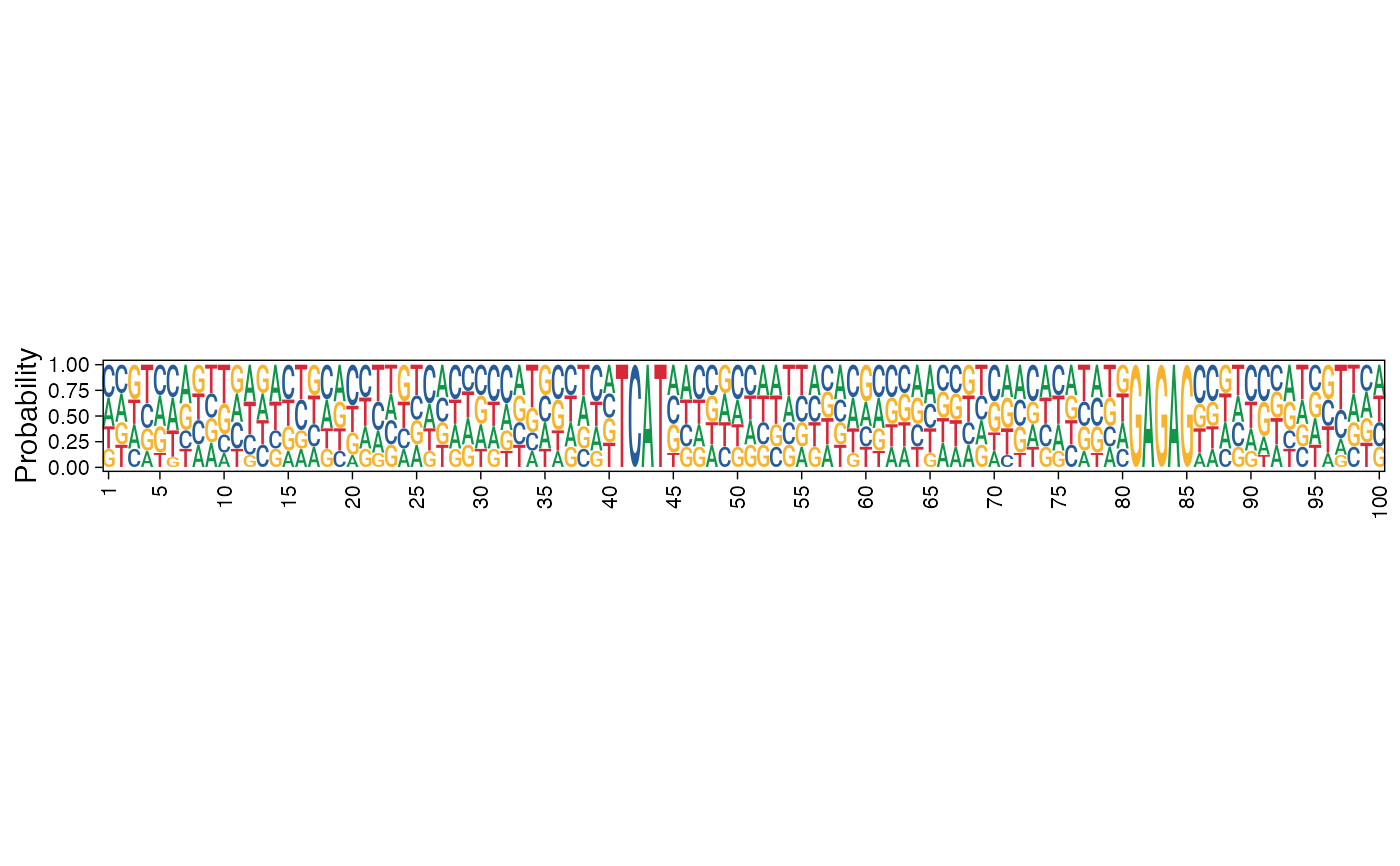
Plot sequence logo of a collection of sequences
Source:R/plot_arch_for_clusters.R
plot_ggseqlogo_of_seqs.RdA wrapper to ggseqlogo plotting. Given a collection of sequences, this function plots the sequence logo.
plot_ggseqlogo_of_seqs( seqs, pos_lab = NULL, xt_freq = 5, method = "bits", title = NULL, bits_yax = "full", fixed_coord = FALSE )
Arguments
| seqs | Collection of sequences as a
|
|---|---|
| pos_lab | Labels for sequence positions, should be of same length as that of the sequences. Default value is NULL, when the positions are labeled from 1 to the length of the sequences. |
| xt_freq | Specify the frequency of the x-axis ticks. |
| method | Specify either 'bits' for information content or 'prob' for probability. |
| title | The title for the plot. Deafult is NULL. |
| bits_yax | Specify 'full' if the information content y-axis limits should be 0-2 or 'auto' for a suitable limit. The 'auto' setting adjusts the y-axis limits according to the maximum information content of the sequence logo. Default is 'full'. |
| fixed_coord | Specify TRUE if the aspect ratio of the plot should be fixed, FALSE otherwise. Default is TRUE. When `method` argument is set to 'bits', ratio is 4, when 'prob', ratio is 6. |
Value
A sequence logo plot of the given DNA sequences.
See also
plot_arch_for_clusters for obtaining multiple
sequence logo plots as a list.
Examples
res <- readRDS(system.file("extdata", "example_archRresult.rds", package = "archR", mustWork = TRUE)) # Default, using information content on y-axis pl <- plot_ggseqlogo_of_seqs(seqs = seqs_str(res, iter=1, cl=3), pos_lab = seq_len(100), title = NULL, fixed_coord = TRUE)#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> #>#>pl# Using probability instead of information content pl <- plot_ggseqlogo_of_seqs(seqs = seqs_str(res, iter=1, cl=3), pos_lab = seq_len(100), title = "", method = "prob", fixed_coord = TRUE)#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> #>#>#>pl