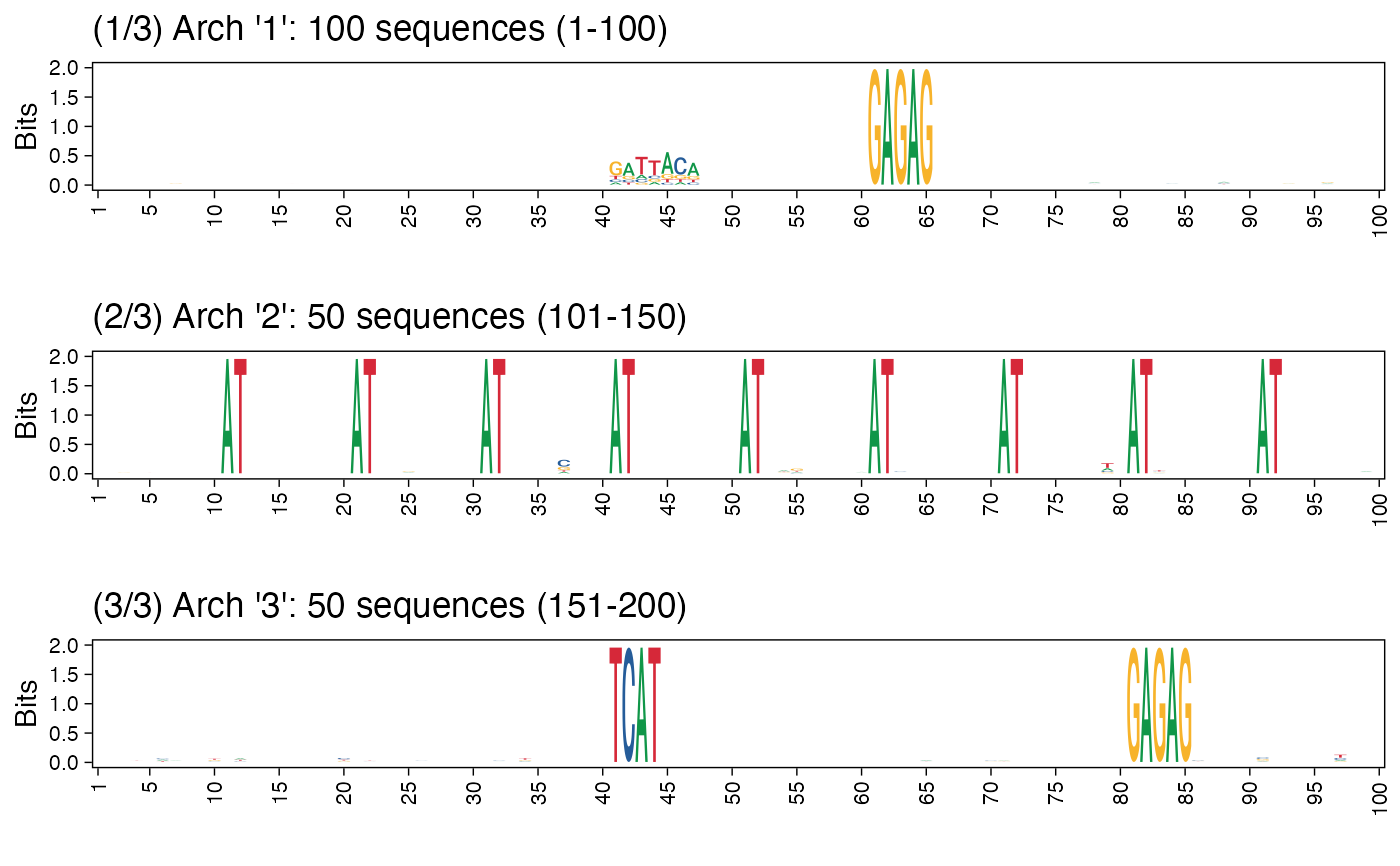
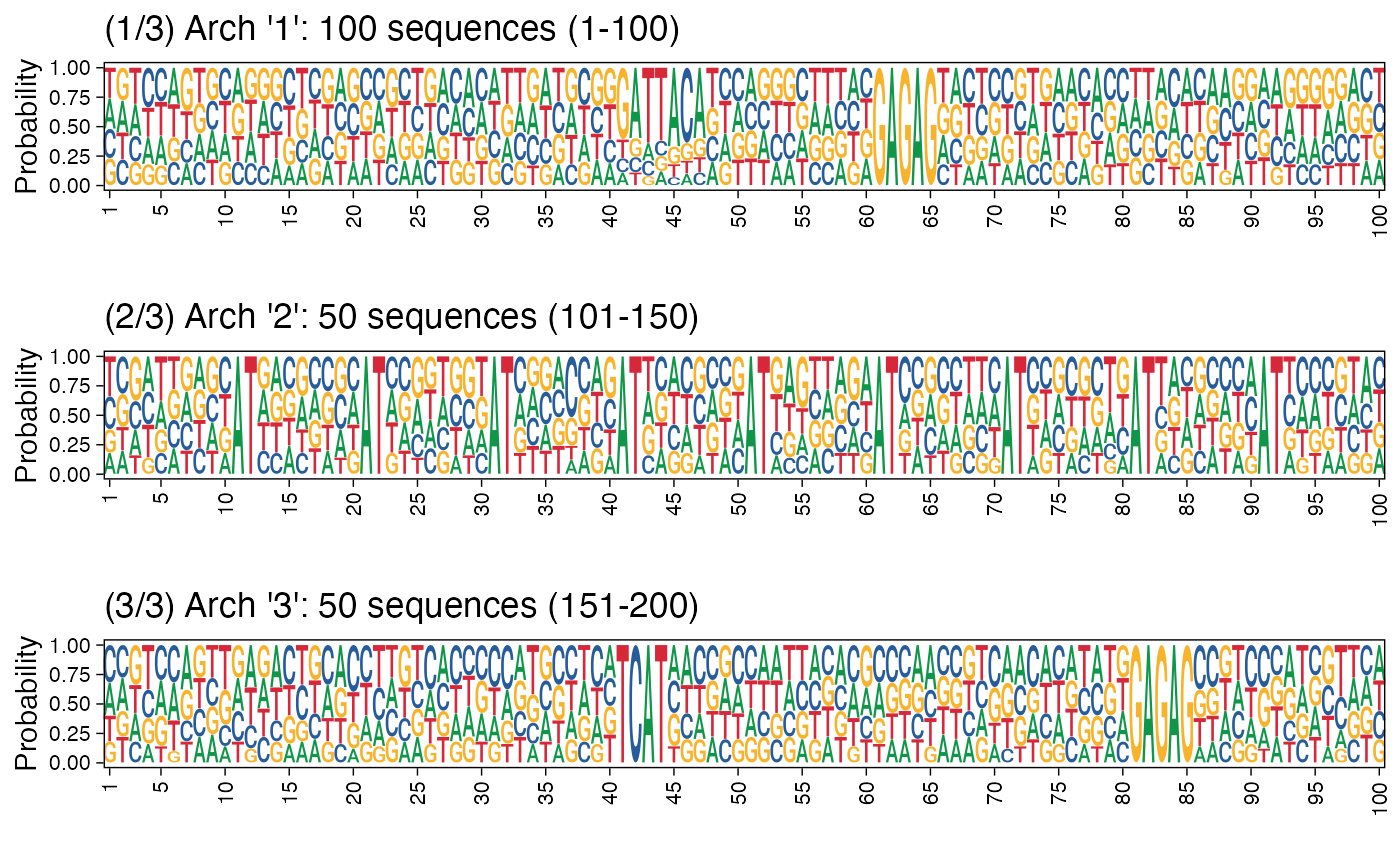
Plot cluster architectures as sequence logos.
Source:R/plot_arch_for_clusters.R
plot_arch_for_clusters.RdGiven a collection of FASTA sequences as a DNAStringSet object, and the clusters information, this function plots the architectures for all clusters. If a name for the PDF file is provided, the resulting set of architecture sequence logos are saved as a multi-page PDF.
plot_arch_for_clusters( seqs, clust_list, pos_lab = NULL, xt_freq = 5, set_titles = TRUE, pdf_width = 11, pdf_height = 2, pdf_name = NULL, show = FALSE, ... )
Arguments
| seqs | Sequences as a |
|---|---|
| clust_list | Clusters as a list of sequence IDs in each cluster. |
| pos_lab | Labels for sequence positions, should be of same length as that of the sequences. Default value is NULL, when the positions are labeled from 1 to the length of the sequences. |
| xt_freq | Frequency of x-axis ticks. |
| set_titles | Specify TRUE if titles are to be written for the plots. With FALSE, there are no titles for the plots. The title for each plot includes the current cluster number, total number of clusters, start and end sequence numbers in the collection. |
| pdf_width, pdf_height | Width and height in inches of the PDF file. Default values are 11 and 2. |
| pdf_name | Specify the PDF filename. |
| show | Set TRUE if plot should be immediately shown/plotted. Default is TRUE. By setting FALSE, one can simply collect the list of plots and use any other approach to arrange/display them. See examples. |
| ... | Additional args passed to |
Value
A list of (ggplot2-based) sequence logo plots is returned. When a valid file name is specified, the list of plots is also written to the PDF file (one plot per page).
Examples
res <- readRDS(system.file("extdata", "example_archRresult.rds", package = "archR", mustWork = TRUE)) # Default position labels 1 to length of the sequences. # Can also set pos_lab based on biology, e.g., use -50 to 49 denoting # 50 basepairs upstream and 49 downstream of the transcription start site # located at position 0. arch_pl <- plot_arch_for_clusters(seqs = seqs_str(res), clust_list = res$clustSol$clusters, pos_lab = NULL, pdf_name = NULL, fixed_coord = TRUE)#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.# Using cowplot::plot_grid arch_pl <- plot_arch_for_clusters(seqs = seqs_str(res), clust_list = res$clustSol$clusters, pos_lab = seq(100), method = "bits", pdf_name = NULL, show = FALSE)#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.# Plotting architecture sequence logos with probability instead of # information content arch_pl <- plot_arch_for_clusters(seqs = seqs_str(res), clust_list = res$clustSol$clusters, pos_lab = seq(100), method = "prob", pdf_name = NULL, show = FALSE)#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.#> Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale> = "none")` instead.